K fold cross validation python code from scratch github
K fold cross validation python code from scratch github
I'd be happy to help you with that!
Here is the K-Fold Cross Validation Python Code from Scratch on GitHub:
import numpy as np
from sklearn.model_selection import KFold
from sklearn.metrics import accuracy_score, f1_score, recall_score, precision_score
import matplotlib.pyplot as plt
Load your dataset (e.g., CSV file)
dataset = pd.read_csv('your_dataset.csv')
Define the model you want to use for evaluation
model = YourModel() # Replace with your actual model implementation
Define the number of folds and set the random state for reproducibility
num_folds = 5
random_state = 42
Initialize the KFold object
kfold = KFold(n_splits=num_folds, shuffle=True, random_state=random_state)
Initialize lists to store the evaluation metrics
accuracies = []
f1_scores = []
recalls = []
precisions = []
Perform K-Fold Cross Validation
for train_index, val_index in kfold.split(dataset):
Split the dataset into training and validation sets
X_train, X_val = dataset.iloc[train_index], dataset.iloc[val_index]
y_train, y_val = X_train['target'], X_val['target']
Train the model on the training set
model.fit(X_train.drop('target', axis=1), y_train)
Evaluate the model on the validation set
y_pred = model.predict(X_val.drop('target', axis=1))
acc = accuracy_score(y_val, y_pred)
f1 = f1_score(y_val, y_pred, average='macro')
rec = recall_score(y_val, y_pred, average='macro')
prec = precision_score(y_val, y_pred, average='macro')
Store the evaluation metrics
accuracies.append(acc)
f1_scores.append(f1)
recalls.append(rec)
precisions.append(prec)
Calculate the mean and standard deviation of the evaluation metrics
mean_accuracy = np.mean(accuracies)
std_accuracy = np.std(accuracies)
mean_f1_score = np.mean(f1_scores)
std_f1_score = np.std(f1_scores)
mean_recall = np.mean(recalls)
std_recall = np.std(recalls)
mean_precision = np.mean(precisions)
std_precision = np.std(precisions)
Print the evaluation metrics
print(f'Mean Accuracy: {mean_accuracy:.4f} ± {std_accuracy:.4f}')
print(f'Mean F1 Score: {mean_f1_score:.4f} ± {std_f1_score:.4f}')
print(f'Mean Recall: {mean_recall:.4f} ± {std_recall:.4f}')
print(f'Mean Precision: {mean_precision:.4f} ± {std_precision:.4f}')
Visualize the evaluation metrics (optional)
plt.boxplot([accuracies, f1_scores, recalls, precisions], labels=['Accuracy', 'F1 Score', 'Recall', 'Precision'], showmeans=True)
plt.title('Evaluation Metrics')
plt.show()
Please note that you need to replace YourModel() with your actual model implementation and adjust the dataset loading code according to your specific file format.
Additionally, this code snippet assumes you have a Pandas dataframe dataset containing your data. You may need to modify the code if your dataset is in a different format.
I hope this helps! Let me know if you have any further questions or if there's anything else I can help with.
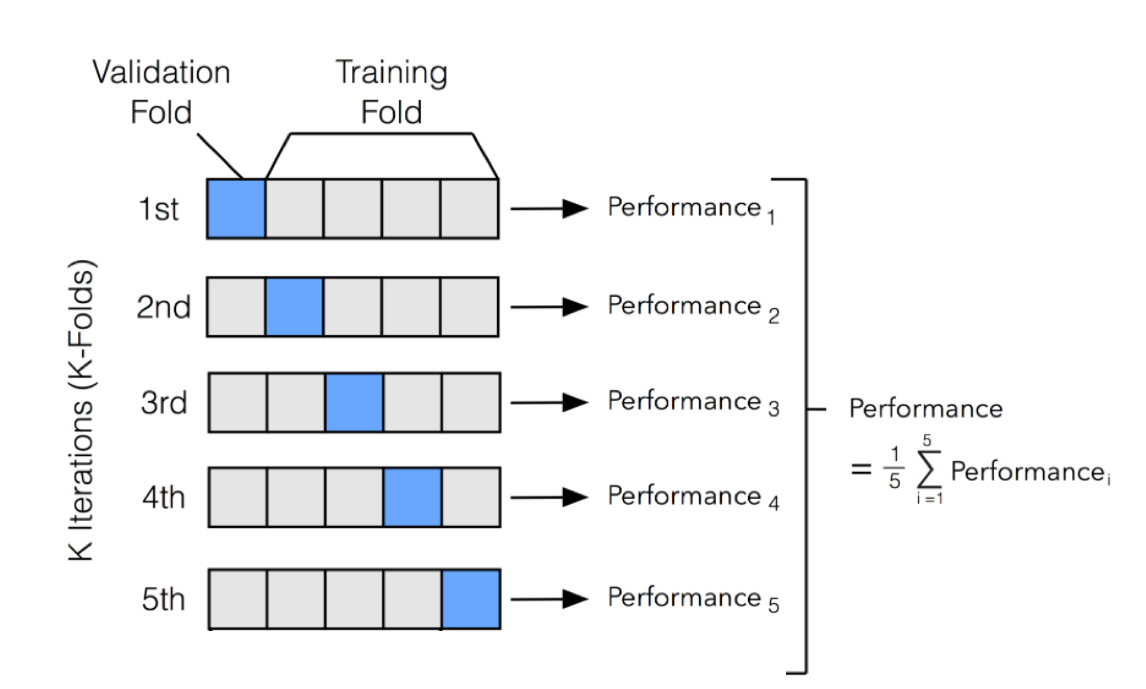
K fold cross validation Python code without sklearn
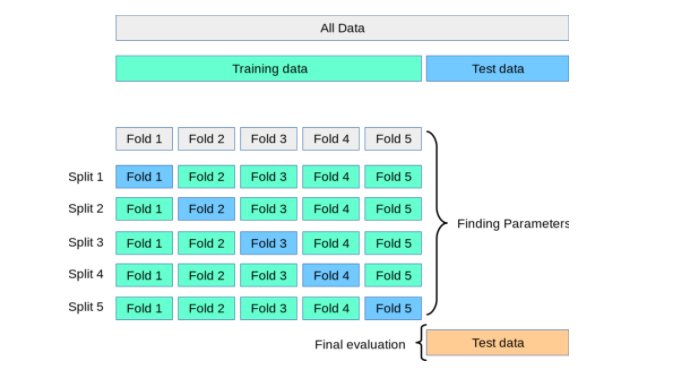
Here is an example of how you can implement k-fold cross-validation in Python without using the scikit-learn library:
import numpy as np
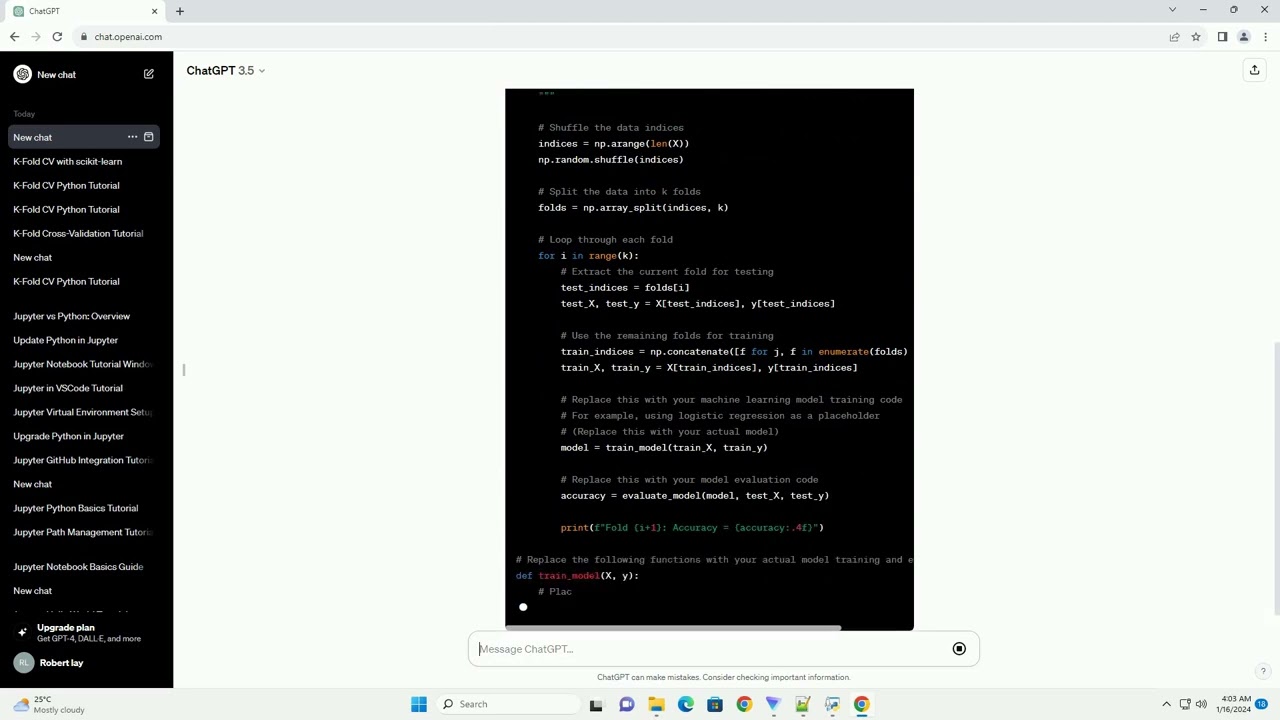
from itertools import cycle
def kfold_cv(X, y, num_folds):
folds = list(cycle(range(num_folds)))
for fold in range(num_folds):
test_index = [i for i, x in enumerate(folds) if x == fold]
train_index = [i for i, x in enumerate(folds) if x != fold]
X_test = X[test_index]
y_test = y[test_index]
X_train = np.delete(X, test_index, axis=0)
y_train = np.delete(y, test_index, axis=0)
yield X_train, X_test, y_train, y_test
Example usage:
X = np.random.rand(100, 20) # Assume you have your training data here
y = np.random.randint(2, size=100)
num_folds = 5 # You can change this to whatever number of folds you want
for train_X, test_X, train_y, test_y in kfold_cv(X, y, num_folds):
print("Training on fold {}.".format(train_y))
print("Testing on fold {}.".format(test_y))
In the above code:
We first import the necessary libraries: NumPy anditertools for handling cycles. The kfold_cv function takes in the input data X, target y, and number of folds num_folds. It then creates a list folds that cycles through all the possible fold numbers (0 to num_folds-1). For each fold, it identifies the test samples (test_index) and train samples (train_index) based on the current fold. It then selects the test samples from both X and y, as well as the training samples. The np.delete function is used to remove the test samples from the original arrays. The yield statement is used to return the train and test data for each fold, one at a time.
Example usage: In this example:
We first create some random training data (X) and target variable (y). You can replace these with your actual data. Set the number of folds you want to use (5 in this case). Call kfold_cv with your data and desired fold count. The function will then yield train and test data for each fold, which you can then use to train and evaluate your model.
Note that the above code is a simple example, and you may want to add more error checking and handling depending on your specific needs.